Regular expressions, or regexs, look at assigned blocks of text to find user defined patterns. For a basic overview of regexs, both how to use regexs and why, see Data Science for Lawyers Lesson 3 – Regular Expressions. Another great site to experiment and test regexs with immediate feedback is regexr, which will highlight regex matches in real time (pay special attention to the sidebar RegEx References for more advanced regex creation).
The grep() family contains in-built R functions that allow us to search strings of text for certain patterns. Of particular importance to us here is the grepl() function. Grepl() scans assigned strings for a regex and outputs TRUE if there is a match and FALSE if there is no match.
One basic example of a regex using the grepl() function:
#REGEX, STRING, and CATEGORYNAME, are placeholders in the code below to show the grepl() syntax. REGEX stands for the regex you use (e.g. "//d+"); STRING is the text you search for a match with the regex (e.g. "Hello World 2020"); CATEGORYNAME is the class or label you assign to a text if the regex found a match.
df$regex<-grepl(“REGEX", “STRING”) #This will generate a list of TRUE/FALSE results for each text string e.g. our article names and append them to a data frame.
df$category<-if(df$regex==TRUE,"CATEGORYNAME") #If true, then this will fill in the appropriate category name in the correct category column of the data frame.
This process can easily categorize a large volume of text given an appropriately precise regex. However, redoing this code for each regex and category we want is inefficient. Repetitive coding like this is not only an eyesore, it also wastes a lot of time, both in the initial time it takes to write (even using copy+paste) and the headache of having to backtrack and edit any regexs or category names! See the GIF below for how quickly this type of coding can get out of hand!
So how do we manage this coding sprawl? Use Loops and the apply() family!
At their most basic, loops simply repeat a function over a vector or data frame. To see basic loops in action, check out the Loops portion of Data Science for Lawyers Lesson 1 – Getting Started. The Apply() family similarly repeats a function, but, unlike a for-loop, does not do so in a particular order, and can provide several different outputs.
But how do we use loops and apply() with regexs?
Say we want to categorize a bunch of text chunks. First, we need to define our categories and design the regex that we will use to find those categories. Then, we can run grepl() using those regexs and save the results to a new value (we will call that category_matches). Let’s try this with an example. We will use regexs to categorize the electronic commerce articles of a treaty by subject matter.
DOWNLOAD the CSV!
This CSV contains a small selection of the ToTA treaty database with actual trade treaty text to test our regexs on. It contains the name of the treaty, year, article names, and article text. We will first categorize the subject matter of an article based on the article name. From previous research we know, for example, that e-commerce treaties deal with consumer protection, transparency and spam.
setwd() #set working directory to appropriate location
df<-read.csv("ToTA_ecommerce_blogsample.csv")
Categories<- c(“Consumer Protection”, “Transparency”, “Unsolicited Commercial Electronic Messages”)
Regexs<- c(“(Consumer|Fraud)”, “Transparency”, “Unsolicited”)
category_matches <- grepl(Regexs, df$article_names) #search for matches (T/F)
Using grepl(), however, will only evaluate the first regex (and give us a logical list of 64 items)!


Our next step could be to write a loop to run grepl() over each regex. Our most straightforward fix here, however, is to use sapply() since it will return us a corresponding matrix (instead of a list) of all the TRUE/FALSE results.
This code will create a matrix (category_matches) by applying the regexs we defined (Regexs) into the function grepl() over all of the article names (df$article_names).
category_matches <- sapply(Regexs, grepl, df$article_names) #search for matches (T/F)
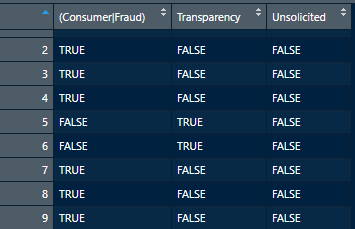
We will also want the column names to reflect the proper category names instead of the regex we used.
colnames(category_matches)<- c(Categories) #rename column names to category names
Next, we will create and add our category column on the original dataframe. We again will use an apply() function, this time with paste() which will take the column name (which is now our category name) whenever the regex found a match. Our new category column will be filled by going down the rows of category_matches and applying the function (u), which here will paste the column name.
df["category"]="" #add empty column to fill
df$category <- apply(category_matches, 1, function(paster) paste(names(which(paster)))) #fill column, “1” tells apply() to go by rows
This apply function looks complicated, so lets break it down and look at the syntax of each part and the variables we used. It is important to remember that apply(), like a loop, works on one rowApply requires three distinct inputs.
- the array you will “apply” over, (category_matches)
- Whether you will apply over rows (1) or columns (2) or both (3), (1 = rows)
- and what function you will “apply”. (function(paster))
But what is function(paster)?
R allows the user to define their own functions (as opposed to built-in functions like sum()). Function() essentially allows the coder to put together a series of functions with quick access to their variables. Functions are helpful where the same series of functions is used but with different input(s). This is often helpful in mathematical calculations, where the formula remains the same but the variable’s values change (see this example of converting Fahrenheit to Celsius).
Our function, paster, is defined within the apply function. Paster consists of three distinct functions:
- Paste – converts vectors to character type and combines them
- Names – gets the names of a set of objects
- Which – returns the location of TRUE values in a logical vector
The series of steps will results in the following: Paster will first note which values are TRUE in category_matches (which indicate a category regex match), it will then take the column name of that match (the category name), and then paste that name onto the corresponding df$category row.
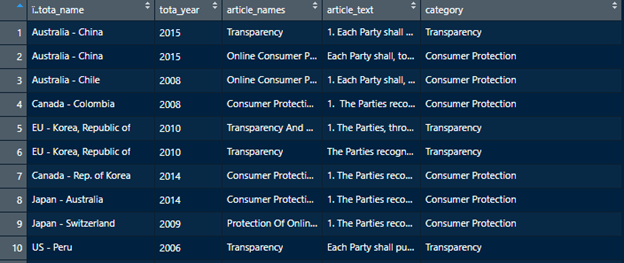
Our dataframe will now be neatly categorized! This will often be all that is necessary for analysis. Sometimes, however, we will want to analyze these categories even further and assign each category its own unique subcategories.
NB: It is entirely possible to use the same method as above and run the regexs for our subcategories over every article in our dataframe again. However, writing regexs that are specific enough to run over the entire dataset without being over-inclusive is excessively difficult. Instead, it would be much easier if we only ran our subcategory regexs on the appropriate category of articles. But how we can we do that while keeping our code compact and easy to manage?
Nested Loops!
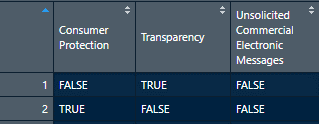
Before we write our loops, we need to make a new data frame that will tell our loops which categories to check for which subcategories with which regex. An easy way to do this is to create and load a CSV. The added benefit of using a CSV is that it is much easier to edit and track your category names, subcategories, and regexs. DOWNLOAD the CSV! (This CSV, as pictured below, is a matrix consisting of our categories, subcategories, and subcategory regexs.)

Load and assign the CSV. The additional argument “allowEscapes=T” tells RStudio to read the regexs as written instead of automatically converting them to plain text. We will also have to add a new column to contain our results again.
subcategory_df<- read.csv("ToTA_Subcategory_blogsample.csv", stringsAsFactors = FALSE, allowEscapes = T, na.strings = "") #load csv with regexs intact
df["subcategory"]="" #add empty column to fill
| [row, column] | Represents |
| df[,4] | article text (the string the regex will inspect) |
| df[,5] | Category column we assigned earlier |
| df[,6] | Empty Subcategory column |
| subcategory_df[,1] | Category name |
| subcategory_df[,2] | Subcategory name |
| subcategory_df[,6] | Subcategory regex |
Next, we create the function that will be used in our loops.
First, we will start with the function that will be looped. We can use the same grepl() function we used above. This time we will use indexes to tell grepl() where to find the proper regex and string to check. The && will ensure our function returns TRUE only if the article is in the appropriate category and the subcategory regex finds a match in the text. This code will return TRUE when the subcategory regex is TRUE and will only run the subcategory regexs on the appropriate categories.
(grepl(subcategory_df[1,1], df[1,5])) && #if "Category" is same, AND
(grepl(subcategory_df[1,3], df[1,4]))) #the appropriate regex is TRUE in the article text
Next, like we did earlier, we can turn that TRUE into a string of our choosing (in this case, the subcategory name). We can do this with an IF statement. IF the entire expression is TRUE, then the code will write the subcategory name to our new subcategory column. If there are multiple subcategories per row (which is highly likely) they will be separated by a semicolon. If the expression is false, nothing will happen.
if
((grepl(subcategory_df[1,1], df[1,5])) & #if "Category" is same, AND
(grepl(subcategory_df[1,3], df[1,4]))) #the appropriate regex is TRUE in the article text
{
df[row,6] <- paste(subcategory_df[1,2], df[1,6], sep = ";")
}
But right now, our code will only work on the first row of our data frame and only look for our first subcategory. We must construct a loop to run that same code on every row of the data frame, and another loop to check for every subcategory.
Our first loop will check every data frame row for the first subcategory. The loop will look from the first to the last rows of our data frame (1:nrow(df)). The value of “row” will start at 1, increasing by 1 for each finished loop, and then stop at the end of the data frame (nrow(df)). We can now use “row” to update our indexes so that the loop will evaluate each row of our dataframe!
rfor (row in 1:nrow(df)) {
if
((grepl(subcategory_df[1,1], df[row,5])) & #if "Category" is same, AND
(grepl(subcategory_df[1,3], df[row,4]))) #the appropriate regex is TRUE in the article text
{
df[row,6] <- paste(subcategory_df[1,2], df[row,6], sep = ";")
}
}
The code above will check the entire data frame for the first subcategory. However, we want to check the entire data frame for every subcategory! We need another loop.
We will assign a new value (row2) that will increase by 1 every time the loop inside it completes (in this case, after each time the entire data frame is evaluated). We need to substitute our new “row2” value for all the subcategory_df indexes. Now, once the first subcategory regex has evaluated the entire data frame, the next subcategory regex will be evaluated, and so on until it is completed.
for (row2 in 1:nrow(subcategory_df)) {
for (row in 1:nrow(df)) {
if
((grepl(subcategory_df[row2,1], df[row,5])) & #if "Category" is same, AND
(grepl(subcategory_df[row2,3], df[row,4]))) #the appropriate regex is TRUE in the article text
{
df[row,6] <- paste(subcategory_df[row2,2], df[row,6], sep = ";")
}
}
}
Our code is complete! Our code now selectively runs our subcategory regexs, allowing us to to write our regexs easier without worrying about being over inclusive and finding false positives in inappropriate articles. We can also easily manage all our regexs, subcategories, and categories from our new CSVs. All within the space of 30 lines!